Tissue morphogenesis is a critical process resulting in diverse organ types within and among species. Tomato offers an excellent system to study fruit morphology because of intense selection pressures implemented by domestication, its tractable genetic history and diversity, and the ease of transformation to test hypotheses. The importance of identifying the factors that shape tomato is underscored by the expectation that this knowledge will be readily transferable to other crops and extend to all higher plants. The approaches we are taking are multifaceted and cross disciplinary, leading to advances in fundamentally important plant growth processes. Ultimately, a better understanding of fruit morphogenesis would lead to insights in plant processes of economic importance, and expedite breeding efforts for varieties that produce vegetables and fruits with dimensions that are required by the market-class.
Robust Organ Patterning by OFPs, TRMs and SUNs in Plant Morphogenesis
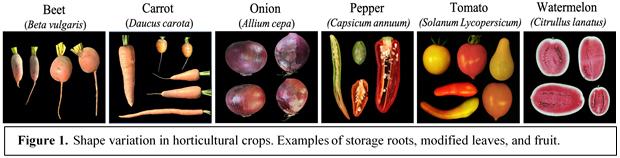
This NSF-PGRP grant is about the recently discovered OFP-TRM-SUN regulon in plants. This regulon has emerged as an important paradigm in the variation in shape of produce ranging from fruits to grains. Proteins from these families are associated with microtubules that are thought to affect cell division leading to altered organ shapes. Still, much remains to be learned about this regulon and whether it extends to root crops. OFP, TRM, and SUN are members of multigene families, yet the role of most members in organ morphology is not well understood. To address these questions, the project plans to map modifiers of tomato sun and ovate in tomato; extend the regulon to carrot and table beet, and investigate the role of SUN in watermelon; discover the function of other members of the OFP, TRM, SUN family; and testing cell growth and geometry parameters by OFP-TRM-SUN. Combined, this information would lead to detailed mechanistic and fundamental insights into the regulation of organogenesis in plants and this knowledge will provide ample opportunities for crop improvement. This project is in collaboration with Irwin Goldman (UW-Madison), Cecilia McGregor (UGA) and Sofia Visa (CoW). For more information: NSF IOS 2048425.
The Role of Ovate Family Proteins in Tomato Fruit Shape
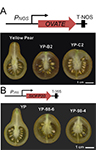
This USDA Agriculture and Food Research Initiative grant aims to discover how OVATE and family members (OFPs) control tomato fruit shape. Factors that determine the successful introduction of crop varieties are the weight and shape of the produce. The appearance often indicates the culinary purpose of the vegetable, whether eaten fresh, sliced, diced or used in stews and sauces. Physical form is also critical for mechanical harvesting, and in general, profitability and market value are positively correlated to larger weights. In recent years, genes that control the shape and weight of the tomato have been identified. These discoveries have enabled targeted breeding efforts to alter or maintain the specific produce dimensions required for the distinct market classes. Despite the knowledge of the genes that control tomato’s appearance, the manner in which they alter the dimensions of the fruit is poorly understood. This has left a gap in our ability to selectively target the morphology of tomato and many other vegetables and fruits. Improved molecular and biochemical insights into the regulation of tomato shape would also increase fundamental knowledge into the formation of plant organs. The focus of this project is to understand how OVATE, a protein that is known to change tomato fruit shape, works together with other proteins to impact cell division patterning. Cell division is a fundamental process in all multicellular plants and animals. Changes in cell division in plants are foundational to larger fruits that have different shapes. For more information: USDA 2017-67013-26199.
Structural variant landscapes in tomato genomes and their role in natural variation, domestication and crop improvement
This National Science Foundation funded project aims to identify structural variants in the tomato genome. A major contributor to trait variation is caused by ‘genome structural variation’ where pieces of DNA are deleted, inserted, or rearranged resulting in changes in gene expression. This project will focus on how structural variation contributed to domestication and breeding of tomatoes. Current whole genome sequencing projects don’t provide a systematic approach to identify structural variation in plant genomes and this is hindering the ability to link genes to important crop phenotypes. This project will unite new long-read sequencing technologies, computational biology, developmental and quantitative genetics, and genome editing to elucidate and manipulate structural variation (SV) at a scale never before achieved for a major crop. By applying SV-detection algorithms to existing short-read Illumina sequencing data from hundreds of accessions, more than 40 genomes will be selected, capturing the majority of predicted SV diversity, to establish new reference genomes using the latest long-read sequencing technology. From these data, a compendium of validated SVs will be generated and integrated with ongoing genome-wide association studies. Significant gene-associated SVs, including those affecting gene activity measured by genome-wide transcript profiling, will be characterized using CRISPR/Cas9 gene editing and quantitative phenotypic analyses, focusing on tomato fruit shape and size that drive crop productivity. This project will greatly expand our knowledge of genomic diversity in tomato, and provide a road map for dissecting SVs in other crops, where such knowledge can be exploited to improve productivity. This project is in collaboration with Zach Lippman (PI, CSHL), Mike Schatz (Johns Hopkins), Joyce Van Eck (Cornell). For more information: NSF IOS 1732253.
Exploitation of Genetic and Epigenetic Variation in the Regulation of Tomato Fruit Quality Traits

This National Science Foundation project aims to use unadapted tomato germplasm for the discovery of fruit quality traits. Wild relatives and semi-domesticated germplasm of cultivated plants provide a significant reservoir of genetic and epigenetic diversity for key regulators of agronomic traits. Future crop improvement relies on harnessing this diversity. However, mining semi-domesticated and wild germplasm for beneficial alleles of agriculturally important traits is not straightforward because fruit quality is quantitatively inherited. Consequently, visual inspection of unselected germplasm does not readily lead to the identification of accessions that have desirable characteristics to improve modern germplasm. The association of traits with genes controlling fruit quality and the identification of beneficial alleles that may have been lost during domestication should provide a model for studying how to efficiently mine germplasm of the closest wild relatives for quantitative trait loci leading to tangible crop improvement. To identify genes and pathways that control complex tomato fruit quality traits, this project will (1) assemble and phenotype a tomato population (Solanum spp.) constituting the continuum of wild, semi-domesticated and ancestral landraces; (2) identify loci underlying fruit quality traits through genome-wide association studies (GWAS) and differentially expressed small RNAs; (3) confirm genetically the traits associated with candidate regions to genes, and (4) analyze the developmental and biochemical pathways that control fruit quality. The project will result in genome sequence data for 150 tomato accessions, including 20 from the closest but fully wild relative of cultivated tomato, 110 from wild and semi-domesticated direct ancestors of domesticated tomatoes and 20 from the earliest landraces of cultivated tomatoes. In addition, the project will generate small RNA sequence data from different stages of tomato fruit development from a subset of this population, and will provide detailed fruit quality information about flavor, firmness, weight and palatability for each of the 150 accessions. The project is in collaboration with Denise Tieman (University of Florida), Ana Caicedo (University of Massachusetts), Lukas Mueller (Boyce Thompson Institute), Sofia Visa (College of Wooster), Jose Blanca, Joaquin Canizares, Maria Jose Diez (Universitat Politecnica De Valencia, Spain). The information will be available through a public resource, the Sol Genomic Network (SGN, http://www.sgn.cornell.edu/), and seeds of the accessions will be available from germplasm repository sites (TGRC, http://tgrc.ucdavis.edu/; and COMAV, http://www.comav.upv.es/). For more information: NSF IOS 154366.
The Regulation and Improvement of Fruit Weight in Fresh Market Tomato
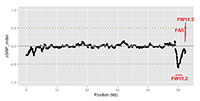
Funded by the United States Department of Agriculture, Agriculture and Food Research Initiative. The molecular regulation of fruit weight among modern tomato varieties is largely unknown despite the importance of this trait in breeding programs. The focus of this project is on five genes that control the weight of the tomato fruit: FW2.2, FW3.2, FW11.3, FAS and LC. Not all of the alleles of the loci controlling fruit weight are fixed in breeding lines and some appear to have negative effects on other agronomically important traits. This implies that crop improvement using alternate alleles for each or all of the fruit weight loci is highly likely. In addition, the identification of additional fruit weight genes would be helpful to enable further characterization of the molecular basis of fruit weight leading to tangible crop improvement strategies. The objectives of this study are: 1) Genotype fruit weight and locule number alleles in contemporary cultivated tomato populations. 2) Develop pre-breeding germplasm for plum, beefsteak and grape market classes, and investigate the utility of known fruit weight alleles. 3) Identify novel fruit weight QTL from populations derived from crosses between underutilized tomato germplasm and those segregating in beefsteak tomato germplasm. 4) Integrate findings with eXtension and publish information on the functional mutations and marker assays for the fruit shape and weight alleles in cultivated tomato, relevant breeding germplasm and commercial hybrids. This project is conducted in collaboration with Samuel F. Hutton, University of Florida; Dilip R. Panthee, North Carolina State University; David M. Francis, Ohio State University. More information about this project can be found at USDA 2013-67013-21229
Funded by the National Science Foundation, Plant Genome Research Program, the main objectives of our tomato fruit morphology grant are i) to identify and characterize genes controlling tomato fruit shape and size, and ii) to discover the underlying molecular networks that control fruit morphogenesis. To do so, transcript, metabolite and hormone profiles will be generated for genes controlling tomato morphology while new shape and size genes will be genetically mapped and cloned. Furthermore, tomato fruit shapes will be described in mathematical terms to aid standardization and classification of the diverse fruit types. Lastly, models of the molecular networks present in tomato connecting gene expression, hormone and metabolite accumulation with morphology will be generated. The co-investigators of this project are: Carmen Catala (Boyce Thompson Institute), Brian McSpadden Gardener (Ohio State/OARDC), Sofia Visa (The College of Wooster). The collaborators of this project are: Carri Gerber (Ohio State/ATI), Yuji Kamiya (RIKEN, Yokohama Japan), Kazuki Saito (RIKEN and Chiba University, Chiba Japan), Simon Gray, John Ramsay (The College of Wooster). More information about this project can be found at NSF IOS 0922661
Transcript profiling of tomato differing in fruit shape and size:
Transcriptome profiling of pooled floral buds (9 days before anthesis and younger) from the SUN-OVATE NILs can be found at http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/experiment.cgi?ID=D005
Transcriptome profiling of various tissues from S. pimpinellifolium LA1589 can be found at http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/experiment.cgi?ID=D006
Transcriptome profiling of young fruit tissues and young flower buds of the FW3.2 NILs can be found at http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/experiment.cgi?ID=D008